In today’s blog, I’ll cover the lastest family of microbiology methods used for testing fuels & fuel associated water. These methods fall under the category genomics – the study of genes. Warning: genetic testing is more technically complex than the methods I’ve described in recent posts. I’ll do my best to keep the language as simple as possible.
Genetic methods have evolved substantially over the past 30 years. They all depend on the polymerase chain reaction (PCR); first reported in 1983. The common steps of all PCR methods include:
1) Genetic material (deoxyribonucleic acid – DNA) extraction (fig 1).
2) Heating to separation of the two strands of DNA’s double helix (fig 2) into two single-strands.
3) Cooling and using an enzyme – polymerase – to convert each single strand back into double stranded DNA (fig 3).
4) Repeating steps 2 and 3 until there are millions of copies of each of the DNA originally extracted in step 1.
5) Using analytical tools to: identify the different types of DNA that were extracted from the original sample in step 1.
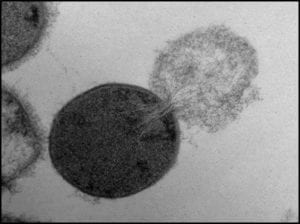
Fig 1. Bacterial cell lysing and ejecting its cytoplasm
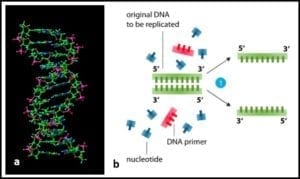
Fig 2. a) DNA molecule ; b) section of DNA being denatured to two single strands.
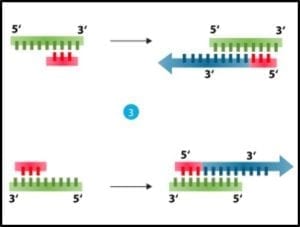
Fig 3. Denatured DNA (left) coupled with primer and reacted with DNA polymerase to form two new double helices.
When PCR methods were first developed in the mid-1980s, DNA was extracted from colonies that had developed on nutrient agar plates (see
Part 12 and fig 4). Early PCR testing revolutionized microbial taxonomy. Microbes that seemed to be closely related because of their appearance and nutrient preferences turned out to be quite distant genetic relatives. Conversely, some microbes that had historically been classified as being members of different groups, were discovered to be nearly identical genetically. However, PCR could only be used to identify microbes that formed colonies.

Fig 4. Bacterial colonies on nutrient agar.
In the 1990s quantitative-PCR (qPCR) methods were developed. In qPCR, messenger-RNA (mRNA) is extracted and used to synthesize complimentary-DNA (cDNA). The PCR process then continues as described in steps 2 through 5. The RNA used for qPCR is typically tagged with a fluorescent dye. A fluorometer is used to measure the DNA concentration as a function of time during repeated cycles of heating and annealing (steps 2 and 3). As shown in fig 5, the time required for the fluorescence to reach a threshold value, can be used to compute the amount of mRNA that was originally extracted from the sample. This, in turn provides an accurate estimate of the population density (i.e., cells/mL) in the sample.
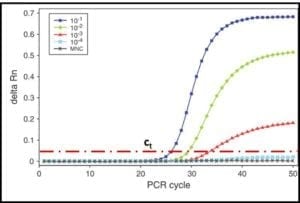
Fig 5. DNA amplification curves: delta Rn is the amount of fluorescence detected and ct is the threshold delta Rn used to compute the DNA concentration in the original sample. The five curves show that the number of PCR cycles needed to reach ct increases as the original DNA concentration decreases.
Four molecules (nucleotides) make up the genetic code (adenosine – A, cytosine – C, guanine – G, and thiamine – T). Each three-nucleotide sequence is the code for a specific amino acid. Thus, long strings of three-letter messages specify the amino acid sequence of enzymes – the cell’s machinery for carrying out all of life’s processes. The total genome of each type of cell (operational taxonomic unit – OTU) is unique. Because each of the four nucleotides – A, C, G, and T – has a unique electrical charge, each OTU’s DNA has a unique net electrical charge. Using a technique called gel electrophoresis, after amplification (step 5) analysts can separate and isolate each type of DNA that was recovered from the original sample (fig 6). They can then sequence the genes and attempt to match the sample’s DNA against a DNA library. The result is a taxonomic profile of the microbes that were present in the original sample.
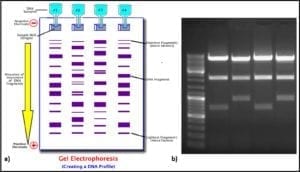
Fig 6. DNA profiling using gel electrophoresis: a) schematic illustration of process ; b) photograph of gel.
In earlier posts, I’ve referred to Donald Rumsfeld’s “unknown unknowns.” Although qPCR and, the more recently variation called next generation sequencing – NGS, is a powerful tool for studying microbial communities in fuel systems, it is probably not the last word in microbiology testing. True, qPCR detects many types of microbes that are undetectable by historically used culture methods. However, extracting DNA or RNA from cells is as much art as science. Genetic material that isn’t extracted isn’t detected. Additionally, qPCR testing depends on the use of primers – short sections of mRNA selected to either be universal (i.e., include a section of A, C, G, T basis that are believed to be present in all bacteria) or specific (i.e., include a section of genetic coding that is unique to a microbe of specific interest). Consequently, researchers are on a steep learning curve about how to select primers. As task force within ASTM D02.14 has just restarted work on a qPCR standard test method for fuels and fuel associated water. The last attempt stalled when participants could not agree on a consensus DNA extraction protocol. As the new task force makes progress I provide readers with updates. The goal is to develop a method that non-technical folks will be able to use.
In the meantime, please contact me at fredp@biodeterioration-control.com you’d like to learn more about fuel system microbiology or microbiological contamination control.
Footnotes:
Source: http://www.newswise.com/images/uploads/2013/01/9/lysis_cover.jpg.
Source: http://www-nmr.cabm.rutgers.edu/photogallery/proteins/gif/dna.gif.
Source: https://laboratoryinfo.com/wp-content/uploads/2015/07/Polymerase_chain_reaction.svg_.png.
Source: https://laboratoryinfo.com/wp-content/uploads/2015/07/Polymerase_chain_reaction.svg_.png.
Source: https://www.microbiologyinpictures.com/bacteria-photos/escherichia-coli-photos/e.-coli-staphylococcu-aureus-colonie.jpg.
Source: https://media.nature.com/full/nature-assets/leu/journal/v17/n6/images/2402922f5.jpg.
Source: http://science.halleyhosting.com/sci/ibbio/biotech/pics/electrophoresisnotes.gif.
Source: https://78.media.tumblr.com/tumblr_lwthmyKO441qzcf71o1_500.gif.







