Since November, this series has progressed through fuel system sampling, sample handling and non-microbiological tests used to detect biodeterioration. This post, and the three to follow, will cover microbiological testing.
Let’s take another look at the figure (fig 1) that accompanied Part 3 (December 2016):
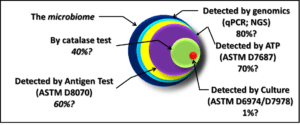
The largest circle represents the total microbiome – all the microbes present in a particular environment. The parameters portrayed in the figure are not exhaustive. For example, the figure does not include direct counts: the use of a microscope to examination samples and count the number of microbial cells per microscope field (the area visible when looking through a microscope’s lenses). Nor does it include tests that measure the concentration of building block molecules such as proteins, carbohydrates, or fatty acid methyl esters (FAME). Direct counting is labor intensive. Moreover, it can be difficult to distinguish between microbes and microbe-size inanimate particles. Finally, there is considerable debate about whether direct counting includes both live and dead cells. Although, theoretically, direct count methods detect 100% of the total microbiome, direct counting is rarely used in practice.
Culture testing is currently the most commonly used tool for determining bioburdens in fuel and fuel associated water samples. Culture testing depends on microbes captured in a sample to be able to reproduce (proliferate) either in or on the growth media used to perform the test. The growth media can be either solid (ASTM Practice D7469), semisolid (ASTM D7978), or liquid (for example: LiquiCult Test Kits – LiquiCult is a trademark of MCE, Inc.; http://www.metalchem.com/liqui-cult.html). First developed in the late 19th century to detect disease causing microbes, culture testing is now often used without any real understanding of its real purpose or its limitations.
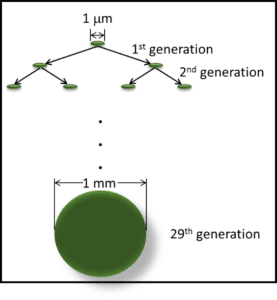
To produce a visible colony (mass of cells), a microbe must reproduce. A generation is the time required for a population to double: for one cell to become two; two to become four, etc. A visible colony has at least 1 billion cells. It takes 29 generations to get from one cell to a billion cells (fig 2).
To reproduce, a microbe must have the right nutrients and environmental conditions. There are more than 5,000 different recipes for microbiological growth media. Each one is optimized for the nutrient requirements of specific types of microbes. No individual type of microbe will grow on all media. Additionally, different microbes have unique preferences for growth conditions (atmosphere with oxygen present versus oxygen-free atmosphere; acidic, neutral, or alkaline environment; cold, temperate, or hot – >40 °C/104 °F; etc.). Consequently, the 1% recovery estimated in fig 1 doesn’t reflect the detection power of all test method combined. It reflects the sensitivity (actually: insensitivity) of any individual culture method. If an analyst ran thousands (perhaps millions) of different combinations of growth media and conditions, the combined results might detect 50% to 60% of the total microbiome population. There are still many microbes that we do not know how to culture.
In addition to the selective effects of any combination of growth medium and incubation conditions, time affects culture test sensitivity. Known microbe generation times range from 15 min for the fastest growing bacteria to 30 days for the slowest. The fastest growing microbes can proliferate from single cells to visible colonies in less than a day. A microbe with a 4h generation time needs nearly five days to from a visible colony, and one with a 30-day generation time needs nearly 2.4 years! Most commercial test kits recommend observing colonies daily, for up to three days. Any microbe with a generation time longer than 2h is unlikely to be detected. Analysts testing samples contaminated with microbes that have generation times of >2h will incorrectly conclude that the samples are uncontaminated.
Notwithstanding these limitations, culture testing has been used with reasonable success for more than a century. It remains the only tool available for obtaining pure cultures on which to do additional testing. Consequently, the take home message is not to dismiss culture testing. Rather it is to recognize that culture testing has specific uses. Obtaining an estimate of total levels of microbial contamination (i.e.: bioburdens) is not one of them. In the next several blog posts, we’ll look at tools better suited for that purpose.

