17TH INTERNATIONAL CONFERENCE ON THE STABILITY, HANDLING AND USE OF LIQUID FUELS

International Association for Stability, Handling and Use of Liquid Fuels (IASH)
IASH was founded in 1983 when Nahum Por, a chemist at Oil Refineries Ltd., Haifa, Israel, obtained support from the Israel Institute of Petroleum & Research to sponsor a conference to discuss issues related to the stability and handling of liquid fuels. The focus was on fuels and crude oil stored as strategic reserves. Between 1983 and 2003, IASH met triennially – venues alternating between Western Europe and North America. Since 2005, the society has been meeting biennially. The association is intimate (approximately 200 members) but include an international group of subject matter experts and people responsible for intermediate to long-term fuel storage. Invariably topics addressed during IASH Conferences range from practical fuel storage and condition monitoring considerations to esoteric explorations of fuel deterioration physicochemical processes. Each conference has included a half-day fuel microbiology session.
I have been attending the IASH conferences since the 1991 Orlando, FL meeting. I have yet to be disappointed by the quality of the presentations and the number of new insights I gain from the speakers. September’s conference in Dresden was no exception.
IASH 2022
The 17th International Conference on the Stability & Handling of Liquid Fuels (IASH 2022) was held in Dresden from 11 through 15 September. In keeping with the precedents set at the previous 16 conferences, the program included excellent papers covering the gamut of fuel quality and stability issues (visit Dresden Agenda (iash.net) to see the conference program). This year’s conference included many papers addressing sustainable fuels – particularly sustainable aviation fuels (SAF) – and fuel quality modelling. I was particularly encouraged to see many younger participants presenting their research. Typically, the conference proceedings are available form the IASH website (Home (iash.net)) three to five months after the meeting. If you are particularly interested in any of the presentations, you can contact the authors directly for copies of their papers or presentations. I won’t attempt to provide a synopsis of every session or paper presented in Dresden. Instead, I’ll focus on the fuel microbiology papers.
IASH 2022 Fuel Microbiology Presentations
Detection Technologies
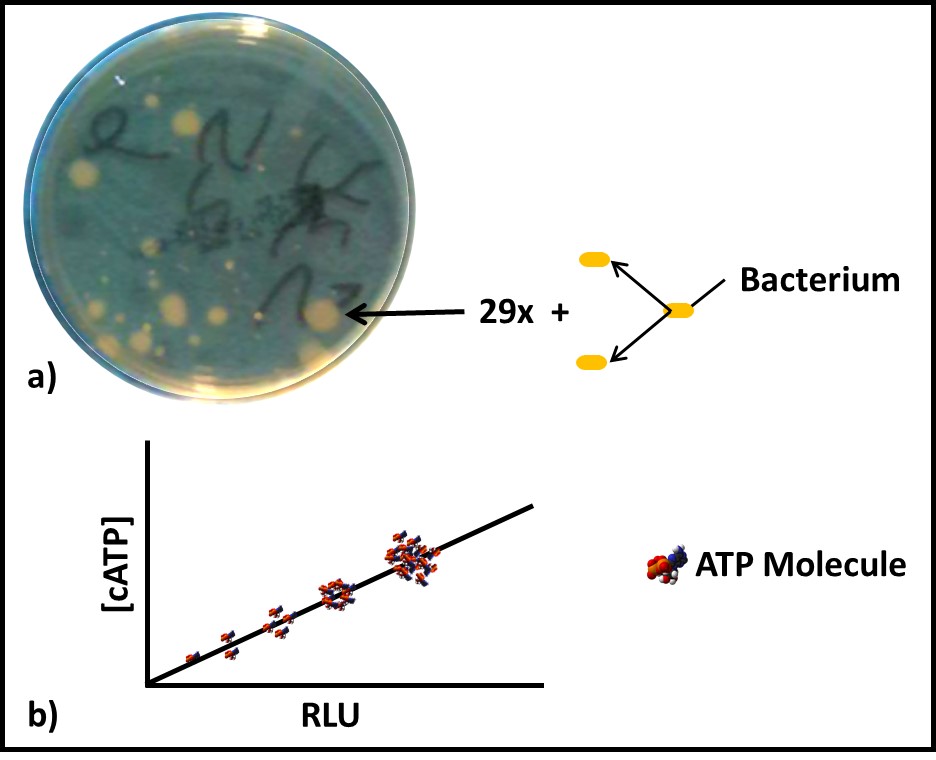
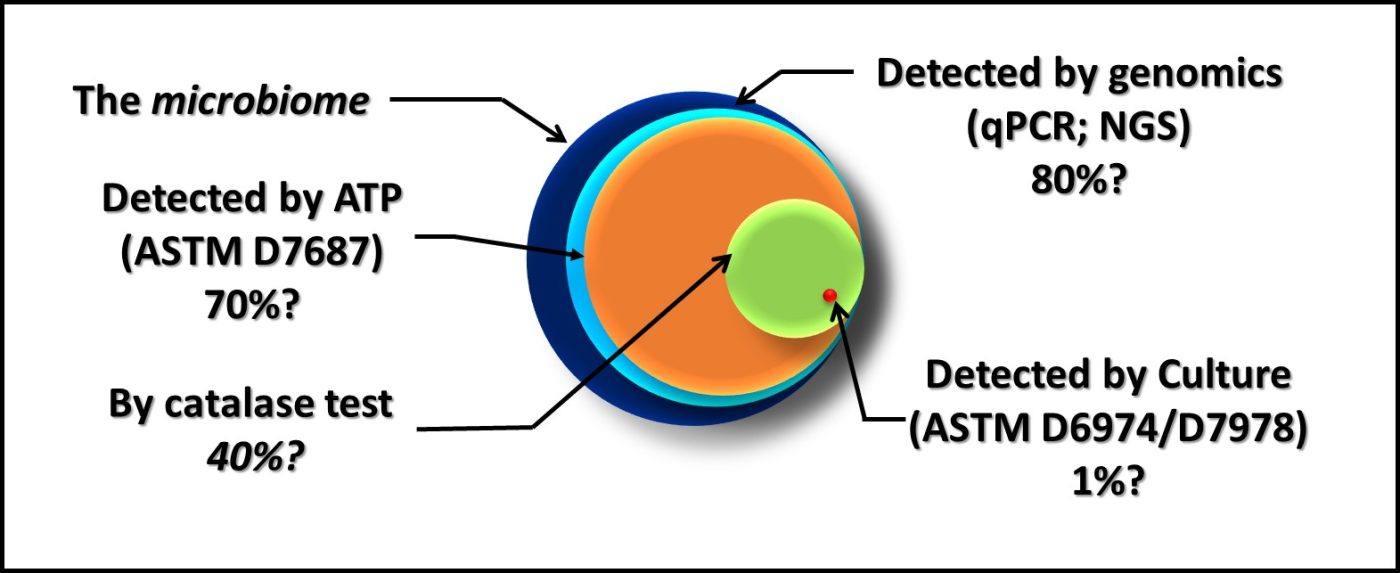
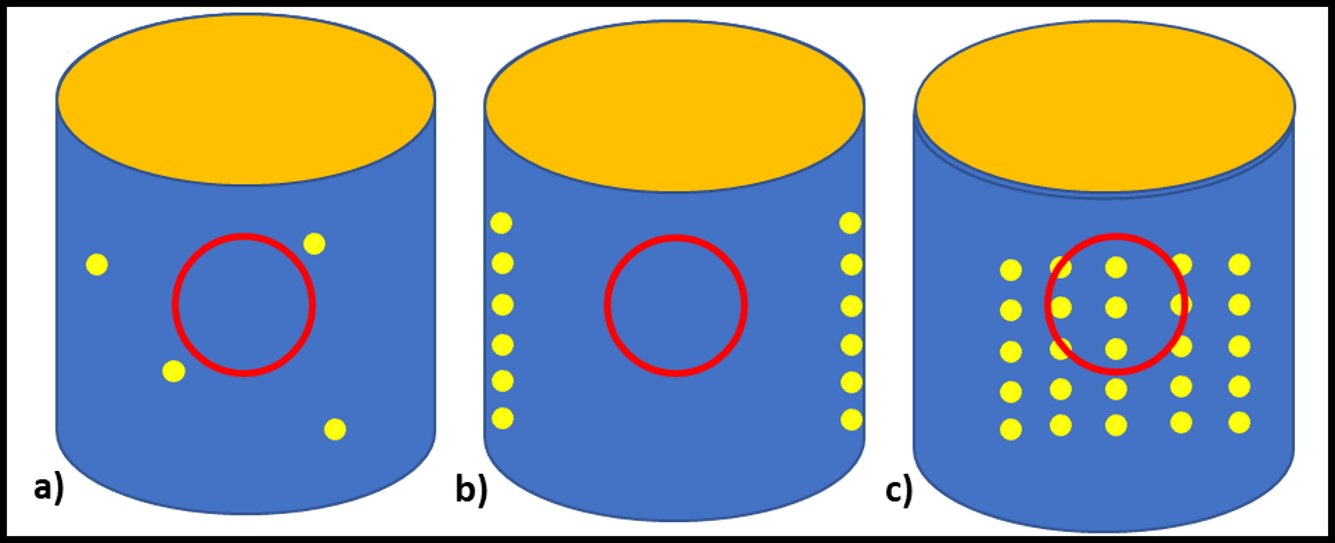
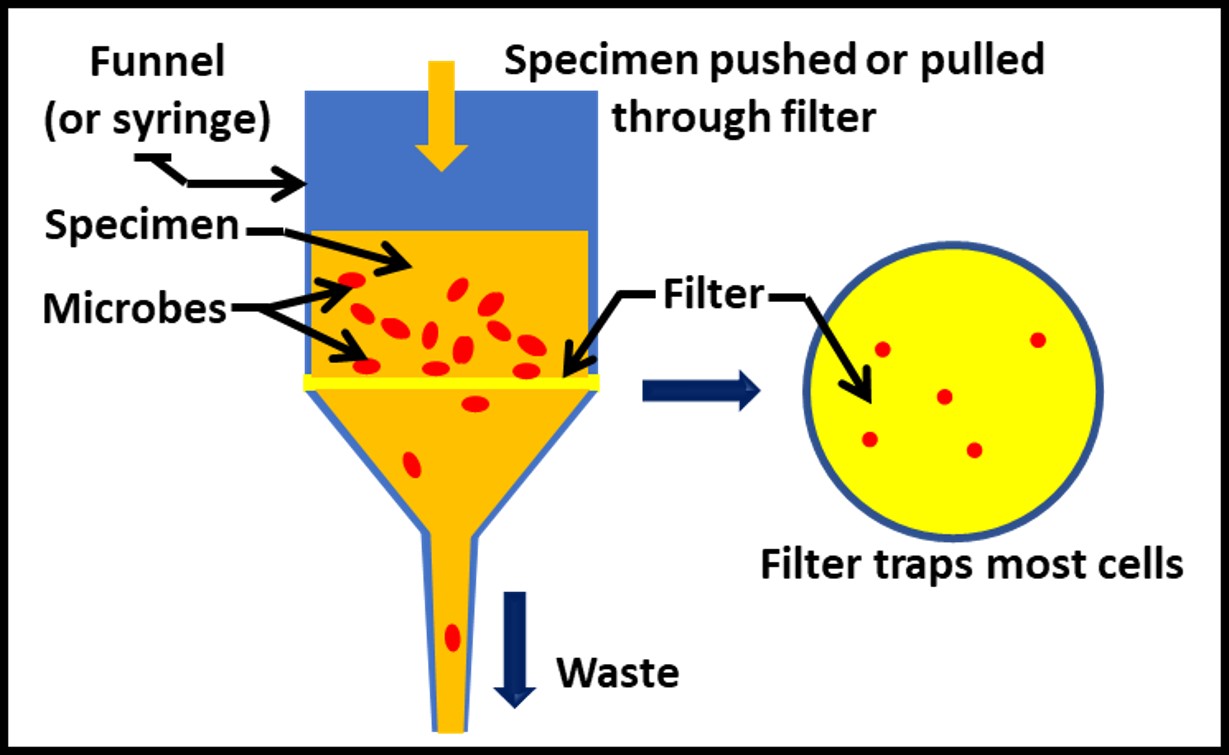
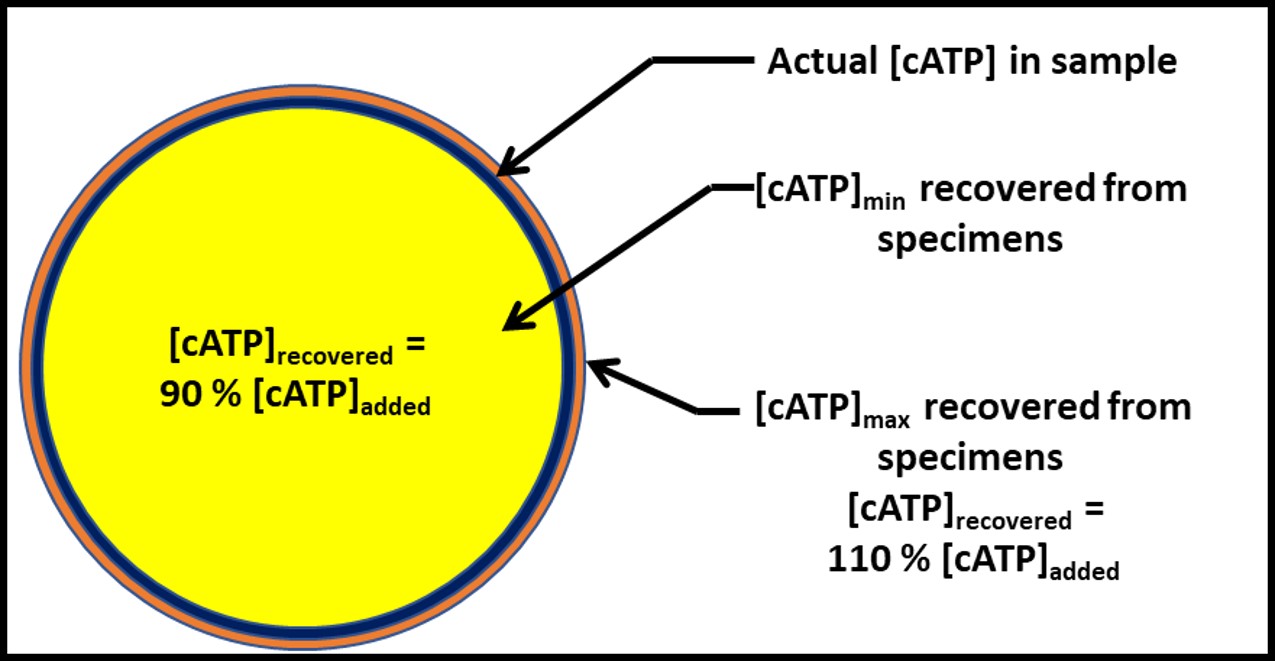
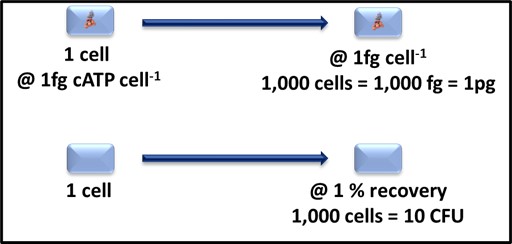
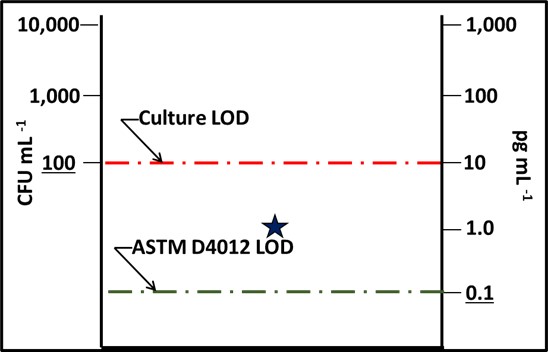
Dr. Jiri Snaidr of Vermicon AG (Hallbergmoos, Germany) reported on the development of a gene probe system for detecting specific microbes in fuel systems. The prototype flow cytometry system relied on fluorescent dye linked, ribonucleic acid (RNA) to label target microbes. After specimens are stained, they are drawn through a flow cytometer. Each target microbe is stained with a dye that fluoresces at a different wavelength so that the abundance of each organism can be quantified. Over the past decade, a new generation of flow cytometer has been developed that has made the technology an increasingly useful tool for quantifying microbial contamination in liquids. There are several challenges related to the use of flow cytometry for microbial contamination in fuels. As I’ll discuss below, the variety of microbes and types that exist in different fuel systems is considerable. Probes designed to detect specific microbes are likely to miss others that might be present. Additionally, microbial population densities in fuel samples is likely to be <1 cells mL-1 . This means that in order to reliably detect fuel contaminant microbes, specimens will need to be ≥1 L.
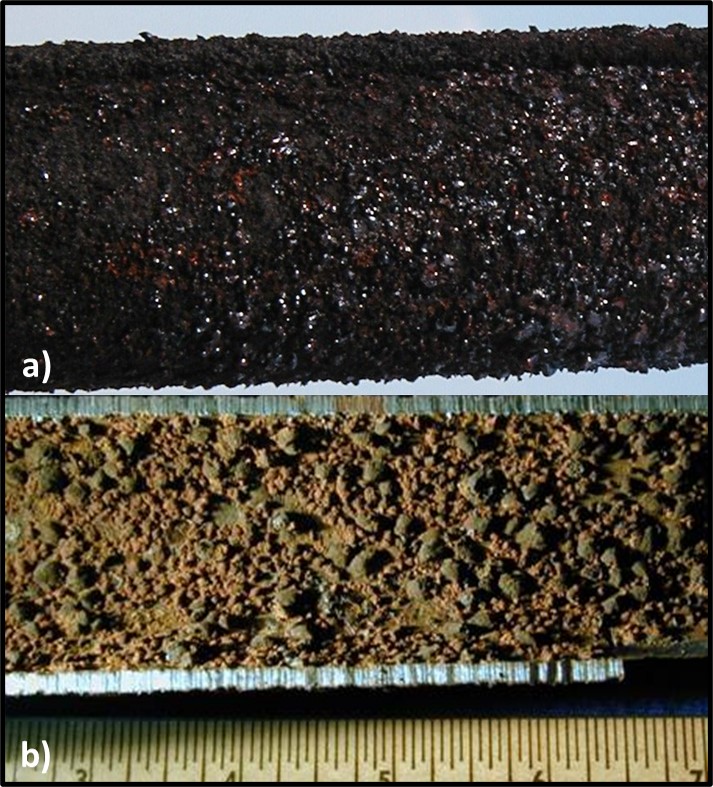
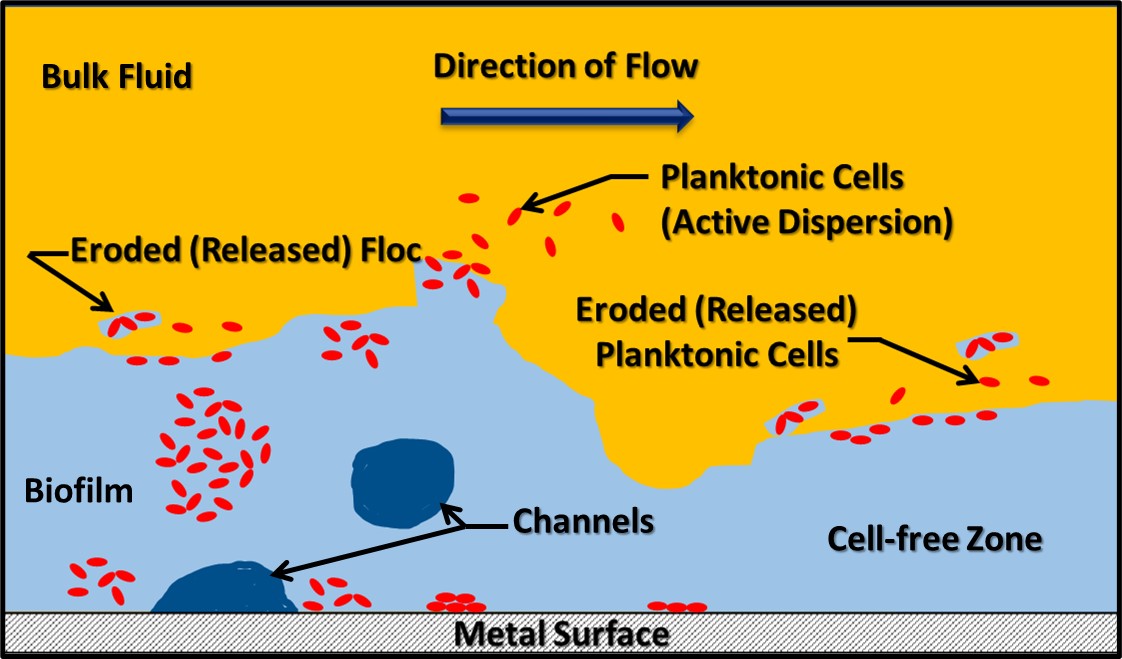
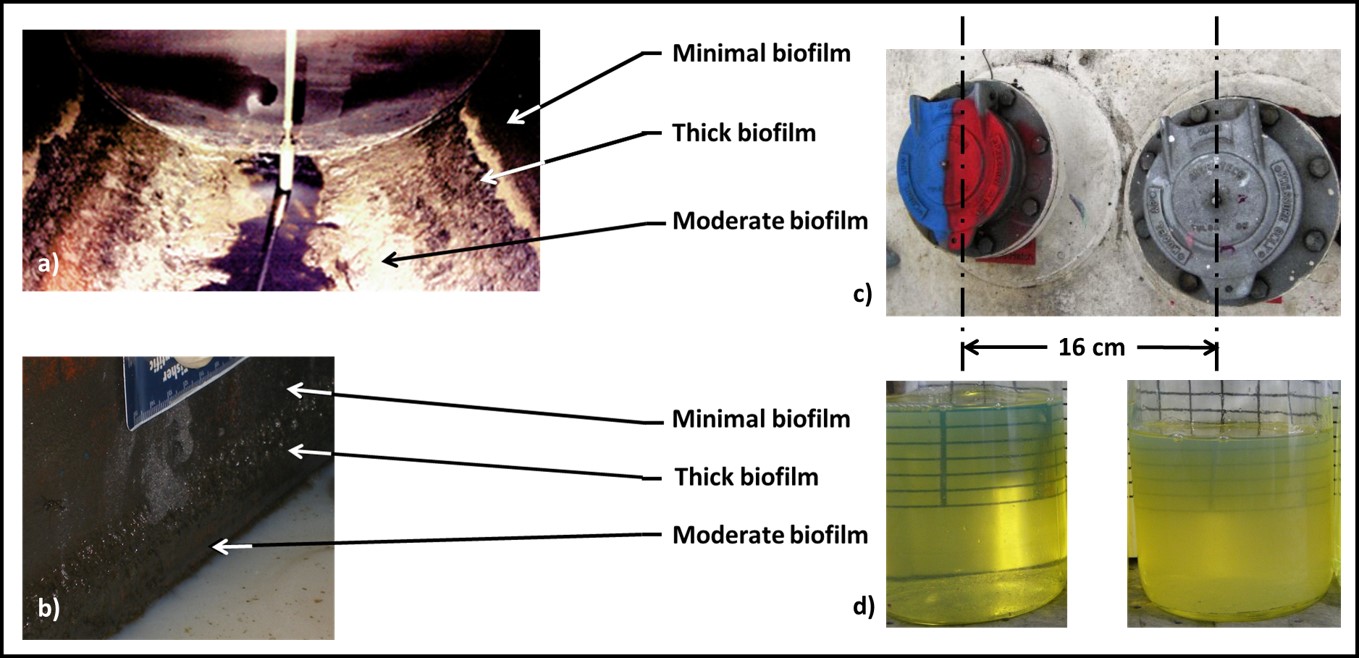
Dr. Osman Radwan of University of Dayton Research Institute (Dayton, OH, USA) discussed the use of an electrochemical biosensor to detect biofilm development on fuel system surfaces. Dr. Radwan’s multi-phase, fluidic device is comprised of an array that is pretreated with a protein molecule believed to be universally present among fuel degrading fungi (i.e., a biorecognition element – BRE). When microbes are bound to the protein, they generate an electrochemical signal. To date, Dr. Radwan and his colleagues have performed in-lab tests demonstrating their ability to detect filamentous fungi and single-cell yeasts. One challenge common among devices such as the one that Dr., Radwan described is that the sensor quickly becomes saturated. The sensors detect initial biofilm formation but cannot differentiate between a 10 µm or 1,000 µm thick film. It is generally recognized that fuel and fuel system biodeterioration are primarily due to the activities of biofilm communities. Thus, being able to assess three-dimensional biofilm growth is an important requirement for surface sensor arrays. Both Dr. Snadir and Dr. Radwan presented cutting edge technologies that are likely to take years of further development before they will be ready for commercial deployment, but their work is exciting and points towards the future.
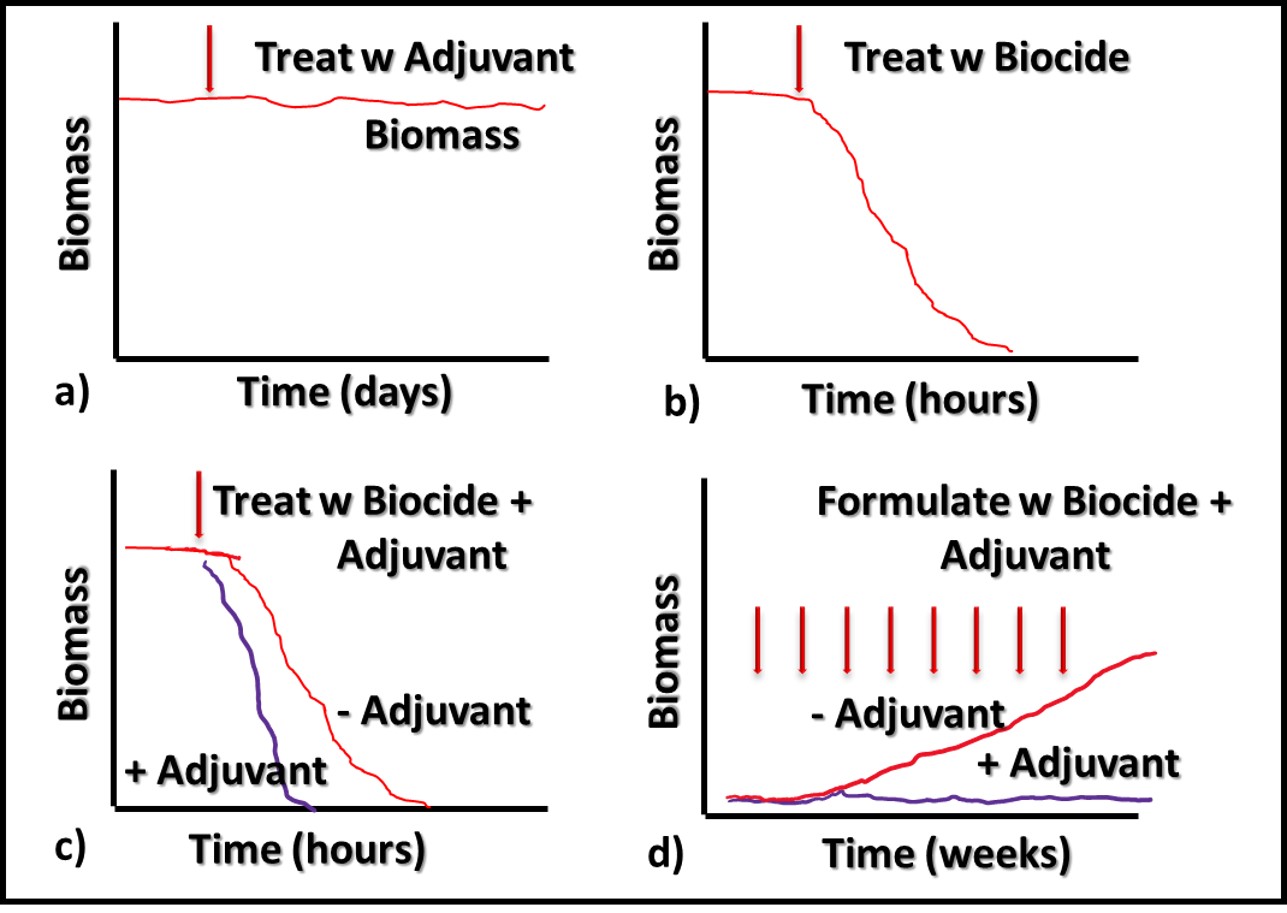
Biodeterioration Mitigation and Contamination Control
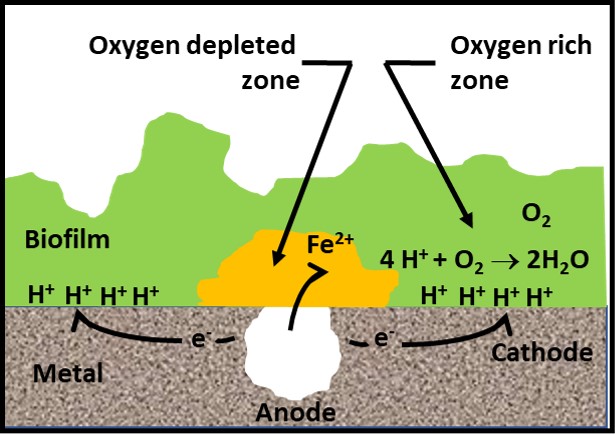
Graham Hill, of ECHA Microbiology, Ltd. (Cardiff, Wales, UK) discussed the relationship between salinity and sulfate reducing bacterial (SRB) activity in marine, seawater ballasted, fuel tanks. Microbiologically influenced corrosion (MIC) has been a problem in ships’ fuel tanks since the early 20th century transition from coal to liquid fuels. In order to maintain stability, ships take on seawater as they consume fuel. One of the most serious, unintended consequences of sea water ballasting is that the combination of seawater and fuel create an excellent environment for the proliferation of microbes. Aerobic and facultatively anaerobic microbes scavenge oxygen and produce low molecular weight organic acids on which SRB can thrive. Mr. Hill reported the results of lab-scale tests run at ECHA. His team found that by using freshwater, rather than seawater – i.e., keeping the salinity at <9 g L-1 (the typical salinity of ocean water is 35 g L-1) – reduces SRB activity (sulfate production) substantially.
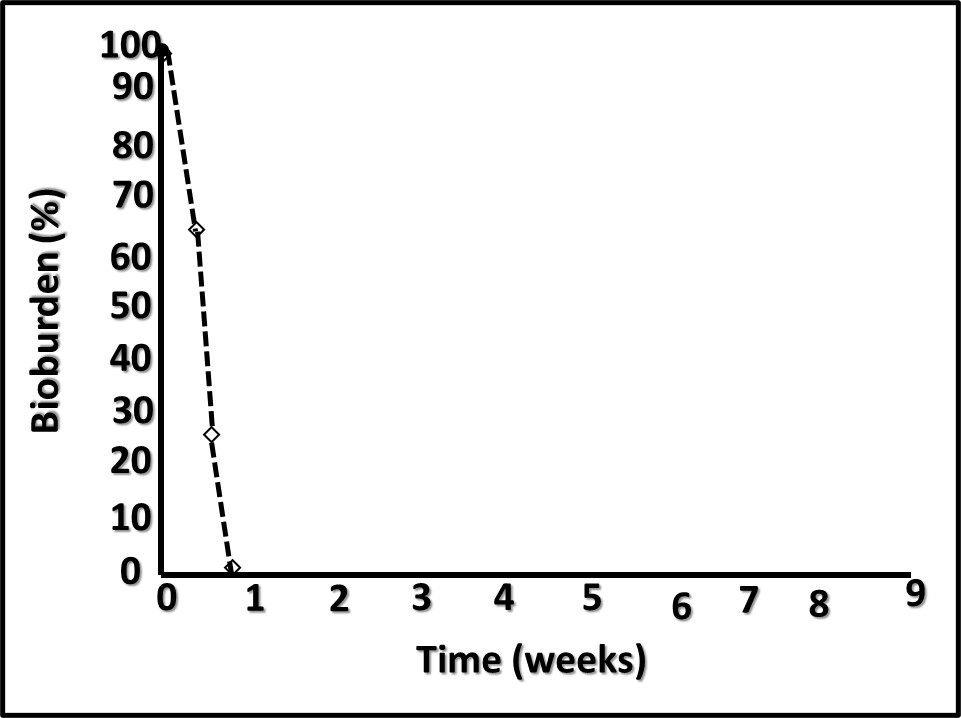
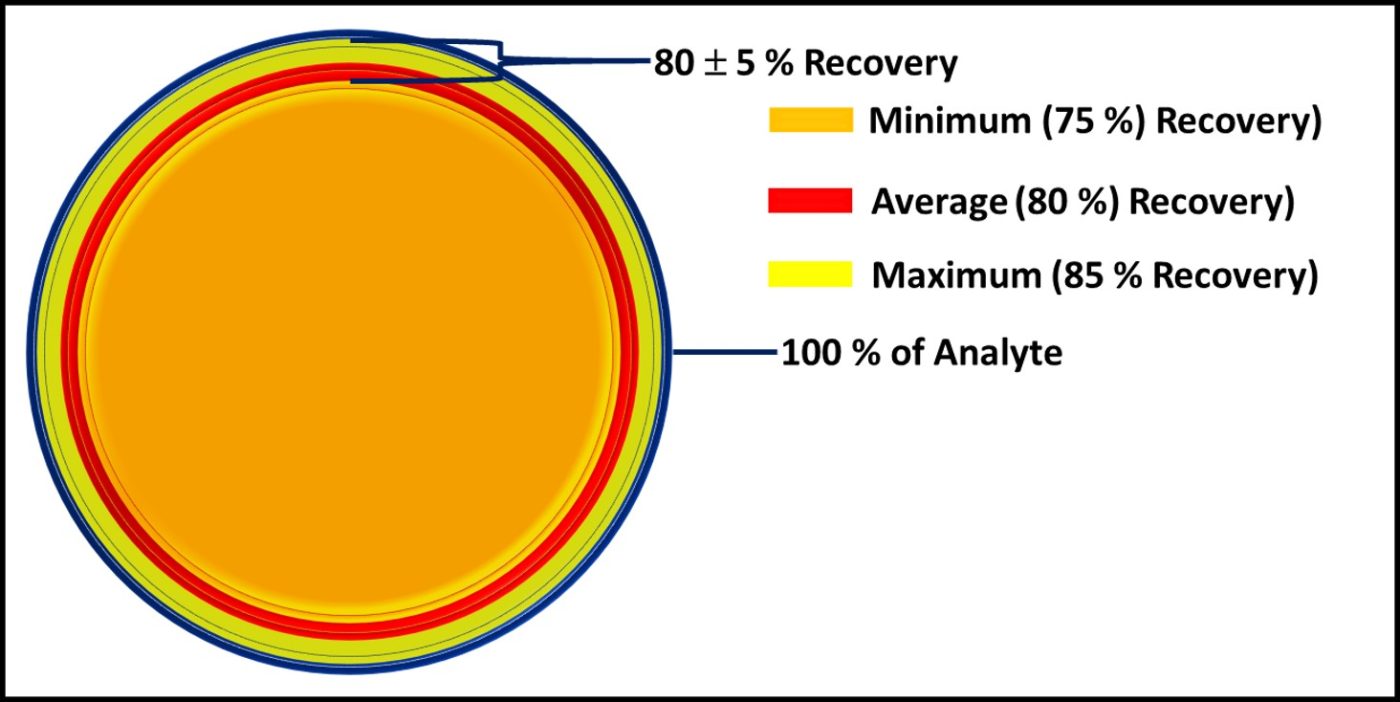
Dr. Oscar Ruiz, U.S. Air Force research Lab (AFRL, Dayton, OH, USA) reported on successful laboratory tests that demonstrated the efficacy of a graphene oxide (GO), nanoparticle, depth filter to scrub microbes from fuel. Dr. Ruiz report the results of a 227 m3 (60,000 gal) trial. At flow rates ≤ 40 L min-1 (≤10 gpm) the device removed >90 % of the bioburden. At the end of the test the pressure differential across the filter was < 28 kPa (4 psi). Given the increased regulatory pressure against the use of fuel-treatment, antimicrobial pesticides (biocides), filtration is a promising alternative. Traditional filter media develop a cake that improves filtration efficacy to a point. Once the filter has accumulated too much material, pressure differentials increase, and flowrates decrease. If GO media can overcome this limitation, they might change the nature of fuel filtration. Filtration’s primary limitation is that the microbes that pass through the medium can colonize downstream surfaces and form biofilm communities. Filtration does not address biofilm control.
Microbial Ecology
Dr. Gareth Wiliams of ECHA Microbiology, Ltd. (Cardiff, Wales, UK) reported the results of an investigation into the factors contributing to hydrogen sulfide generation in salt caverns in which butane was stored. The ECHA team used quantitative polymerase chain reaction (qPCR) and next generation sequencing (NGS) to identify microbes in salt dome samples. Dr. Williams reported that of three caverns tested, hydrogen sulfide accumulated only in one. However, the microbial population profiles were similar among all three caverns. All three had SRB and Halobacterium species (Halobacterium is a genus within the domain of Archaea). The primary difference among the three caverns was the suction line installation. The cavern in which substantial concentrations of hydrogen sulfide had developed had an irregular floor. The uptake line was above the bottom; allowing brine to accumulate. The uptake lines in the other two caverns were at the bottom. In these caverns there was no bottom brine layer.
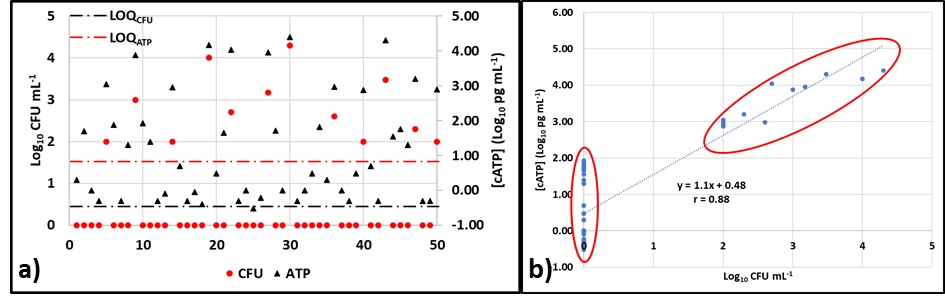
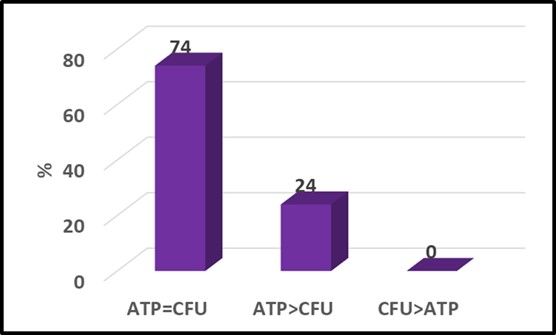
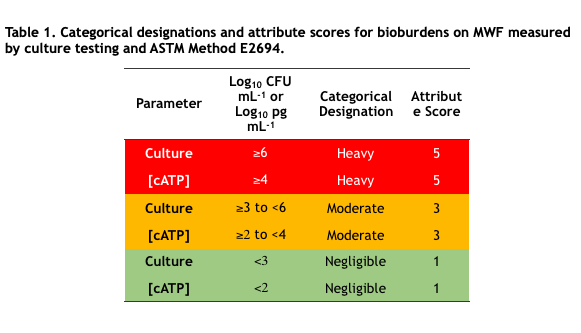
I presented NGS genomic data from the CRC-sponsored diesel fuel microcosm study from which I had reported adenosine triphosphate (ATP and adenylate energy charge (AEC) results at ICSHLF 16 in 2019 (The Relationship Between Planktonic and Sessile Mirobial Population Adenosine Triphosphate Bioburdens in Diesel Fuel Microcosms | IASH Online Library of Conference Proceedings and Newsletters (conferencespot.org) and The Relationship Microbial Community Vitality and ATP Bioburden in Bottoms Waters Under Fuel Microcosms | IASH Online Library of Conference Proceedings and Newsletters (conferencespot.org) – both are accessible to IASH members only. If you cannot access the papers, contact me for copies). Although during the original study (DP-07-16-01-FINAL-REPORT-REV-30JUL21-COMPLETE.pdf (crcao.org)) only a few aqueous phase samples were subjected to NGS testing. We were able to obtain the microcosms after 18-months. Marathon Petroleum, LuminUltra Technologies Ltd., and BCA collaborated to collect samples from the fuel-water interface and aqueous phases f 32 microcosms. LuminUltra Technologies Ltd. Performed the 18-month NGS tests and a different lab had performed the 3-month tests. Among the eight microcosms for which there were data from 3 and 18 months, the genomic profiles of four microcosms had remained stable and those of the other four changed substantially. Duplicate samples from several 18-month microcosms demonstrated that variability among duplicate aqueous samples was excellent but that variability among duplicate interface samples was substantial. Genomic test methods continue to evolve rapidly. As the protocols are refined, it will be essential to understand the sources of variation related to sampling and testing. Without this understanding, results from environmental samples will be difficult to interpret. I’ll devote a future What’s new post to genomics, transcriptomics, and metabolomics.
Lifetime Achievement Award
During the conference banquet I was taken totally by surprise when I was called to the stage to receive IASH’s Lifetime Achievement Award. I confess that when 1 st Vice-Chair, Matt DeWitt announced that it was time to present awards, I commented to my tablemates that these awards have never been given to microbiologists we are such a small minority within the Association. As I was making the comment, I heard my name. Did I mention that I was taken totally by surprise?
The award citation reads:
“By the members of IASH in recognition of his significant technical contributions and leadership related to testing, control and mitigation of microbiological contamination in fuels and oils. His notable technical contributions include over 70 technical publications on fuel, lubricant and metalworking fluid microbiology and biodeterioration. He has had numerous Chair and leadership positions in ASTM, IBBS, and IATA, and contributed significantly to the development of best industry practice documents and guidelines for the prevention and control of microbiological contamination in fuels.”

1st Vice-Chair Matt DeWitt presenting IASH Lifetime Achievement Award to Fred Passman
Summary
In summary, the conference was an exceptional opportunity to learn about various aspects of fuel degradability and its control. I most strongly recommend that individuals who are responsible for fuel quality join IASH and make plans to attend future IASH conferences. Again, for more information about IASH, visit Home (iash.net).
To contact me, please send an email to fredp@biodeterioration-control.com.